Model development
from terascale to petascale computing
My main workhorse is propag, a code for the simulation of cardiac electrophysiology. Originally designed to run on shared-memory systems with a moderate number of processors (32 to 64 at the time), it was initially parallellized with OpenMP. At the Institute of Computational Science in Lugano, the code has been upgraded to a "hybrid" MPI-OpenMP parallellization. The MPI parallellization allows it to run on distributed-memory computers with thousands of processors, while the OpenMP inner loops allow it to be more efficient on systems with "fat nodes", in which dozens of compute cores share their memory. The upgrade of propag has been mainly the work of Dorian Krause.
The code scales excellently to tens of thousands of cores. For most practical applications performance is i/o limited, so production runs are typically performed on a modest 300 nodes (9600 cores) of Monte Rosa, a Cray XE6 computer at the Swiss National Supercomputing Center CSCS.

reaction-diffusion models of the heart
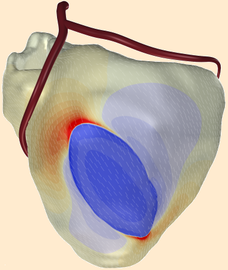
The first serious heart model that I have been involved with was that developed by Marie-Claude Trudel, directed by Ramesh Gulrajani. It worked with 12 million points at a resolution of 0.25 mm [1]. It required 12 GB memory to work, necessitating the use of a supercomputer, then an SGI Origin 2000 (Trudel et al, 2004). Later work (actually published earlier, in 2003) reduced memory consumption to less than 5 GB, making it possible to run the model on a high-end PC [2]. Still, this would require several days computation time. Running on a supercomputer with 16 processors, the model could simulate a full heart beat in 5 hours.
With more realistic membrane models, memory usage quickly goes up, and the perspective of running on a desktop PC becomes more remote [3].
| [1] | Trudel et al. IEEE Trans Biomed Eng 2004 |
| [2] | Potse, Dubé and Gulrajani, Conf IEEE-EMBS 2003 |
| [3] | Potse et al, 34th Int Con Electrocardiol, June 2007 |
bidomain models
Bidomain models are much harder to solve than monodomain models because they require the solution of a system of linear equations, one equation for each model point, at each time step. Such systems are more difficult to solve when they grow larger. For a human heart, tens of millions of points are necessary. In December 2006 we published the first such model. With this model we have shown that, for ECG simulation, a monodomain model is accurate enough. This is good news for researchers who don't have access to supercomputers: a monodomain model of the human heart can in principle run on a (high-end) PC. Bidomain models remain necessary for the simulation of, for example, defibrillation shocks, and for the simulation of extracellular potentials in the heart. For the latter, however, a shortcut can be taken: propagation of action potentials can be simulated with a monodomain model, and from the results extracellular potentials can be computed at relatively large intervals. This still requires a bidomain model and a supercomputer, but it is about 3 times faster than solving the propagation problem with a bidomain reaction-diffusion model.
In October 2007 we have successfully tested our software with a bidomain model of 2 billion (2,000,000,000) points. This is a severe test, because the program must solve a system of 2 billion linear equations for each time step. It required an entire 768-processor Altix 4700 computer, with 1.5 terabyte core memory. Such huge bidomain models may, in a few years from now, become useful to do whole-heart simulations with details as small as a single cell. This will allow a much more realistic representation of the complex structure of the cardiac muscle.
| [1] | Potse et al., IEEE Trans Biomed Eng 2006 |